UnknownProcessor is used by several labs for processing GCMS Scan data for unknown compounds. It can be used for perfume analysis, emission analysis, etc.
UnknownProcessor has been rewritten from scratch and improved on many sides. It is now a super high productivity and standalone tool. It handles 200+ samples with 200+ compounds with ease. Corresponding compounds are lined-up across multiple samples. And that makes reviewing so much more productive.
Some of the function are: Deconvolution, integration, FID and GCMS peak correlation, Retention Index, multi-sample compound lineup, export to Excel, reporting with Excel templates, Toluene equivalent quantitation, FID with GCMS formula quantitation.
Spectrum extraction, library search, FID correlation and Lineup are all done automatic. Your manager will be blown away with the speed of processing from injection to reporting.
An ESTD response/amount or an ISTD (Toluene or another compound) can be used to do semi-quantitation on FID or TIC response.
Corresponding compounds have the same colors througout the application
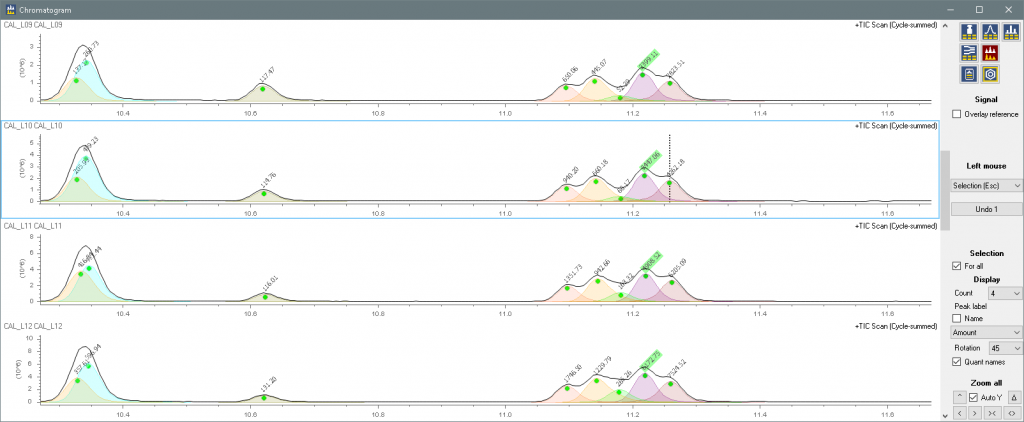
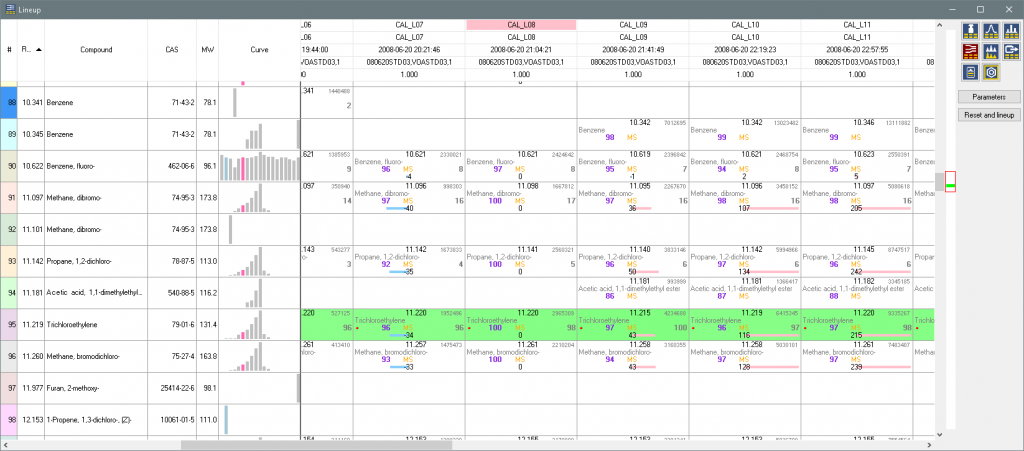
And can be viewed in the lineup table.
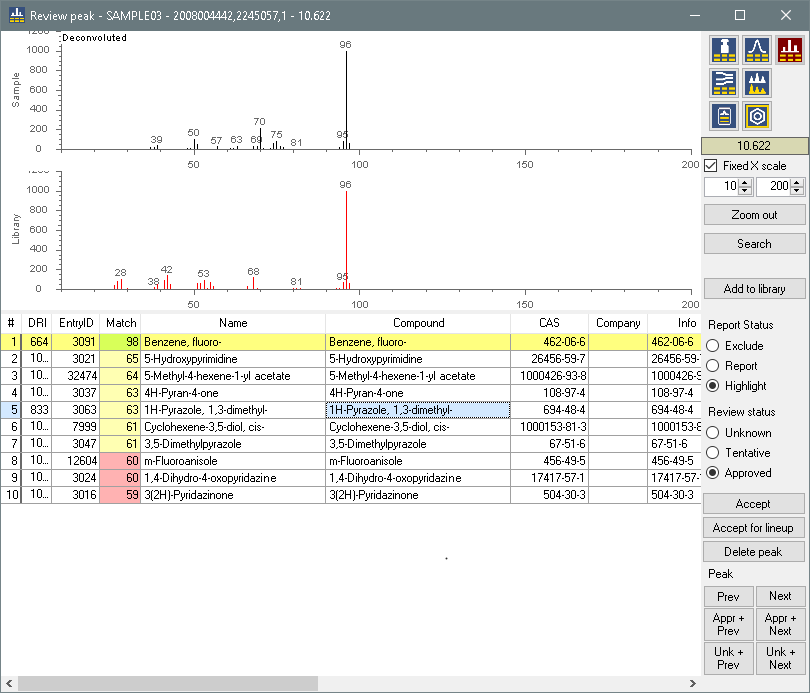
This is the hit view, the tool where you quickly can review the indentifications and move to the next compound.
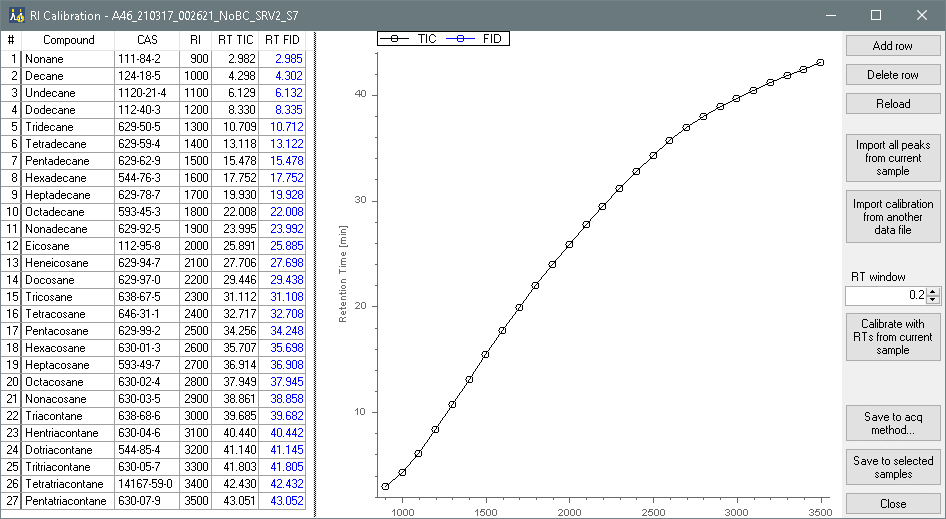
And there is a RetentionIndex tool
Several report templates are available. And you can easily modify report templates in Excel.
There is much more. When you are interested, send an Email and we will do a remote demonstration.
MassHunter add-on
Unknown processor relies on MassHunter Qualitative Analysis. A valid license from Agilent is required.
Pricing
Single instrument license 8000 euro.
Downloads
PDF training
UnknownProcessor Demo version 1.50 setup
History
| Date | Version | Fixed/Changed |
|---|---|---|
| 2024-04-02 | 1.17 | ReviewStatus chrom peak dots did not update Library Limits seems not to work Lineup automation does not work after deconvolution Spectrum window anchor plot Resp% in Lineup Export infinite |
| 2024-06-20 | 1.18 | Removed unused integration parameters from the method Fixed bugs in Lineup and retain lineup after loading batch |
| 2024-06-26 | 1.19 | Option to exclude hits when RI match fails. Option to hide unidentified lineups |
| 2024-09-09 | 1.20 | Improved the IonChromatogram window, show reference sample |
| 2024-09-10 | 1.21 | Improved the Spectrum window, show reference sample |
| 2024-09-25 | 1.22 | Add Hit window NIST search button |
| 2024-10-01 | 1.23 | Option to filter not identified peaks in Peak and Lineup view. Lineup view, added Average amount column. Sample view, added Sum Amount column. Library settings, Use max dRI title. |
| 2024-10-04 | 1.24 | Don’t deconvolute on non-selected chromatograms. Much improved spectrum view (show multiple peaks and lineups). |
| 2024-10-08 | 1.25 | Sample group column Labels of Spectrum View and EIC view Plot.cs crash: SetScaleLimit Overlay ref, check with one sample: crash if no ref sample present Set WellKnown window near to cursor Fix Set Series, names (!Alkane), CMP Group wrong Add to library Tentative: appends constantly SpecWindow Right margin increase Deconvolution should work only on the selected chromatogram Added feature for 2 amount level limits in spectra library and peak table Fixed crash on Extracted Ion view with only one mass |
| 2024-10-08 | 1.26 | Revert deconvolution setting to version 1.21 Peak.Name used for adding to library and RI Calibration instead of Peak.Compound |
| 2024-10-21 | 1.27 | Sample group amount average and RSD% |
| 2024-10-29 | 1.28 | Fixed Sample groups amount average and RSD% After crash on missing last used data path, fall back to c:\ |
| 2024-10-30 | 1.29 | Reorganized the menus Add mass signal multiplier to correct deconvolution errors: Lineup and Chromatogram views: context menu: Calculate Multiplier |
| 2024-11-13 | 1.30 | Lineup Calculate Multiplier for Deconvolution correction on multiple lineups |
| 2024-11-19 | 1.31 | Improved spectrum window |
| 2024-11-01 | 1.32 | Do not reset Peak ReviewStatus after Accept in Hit Review |
| 2024-11-27 | 1.33 | Add logbook exception on deconvolution error |
| 2024-11-28 | 1.34 | Chromatogram view, added the missing menu item: Delete all selected peaks |
| 2025-01-27 | 1.35 | Improved interaction between chromatogra, spectrum and hit view |
| 2025-01-30 | 1.36 | Chromatogram view, new button: copy to clipboard |
| 2025-01-30 | 1.37 | Fix Spectrum view auto update |
| 2025-02-12 | 1.38 | Allow merging of mixed integrated and deconvoluted peaks |
| 2025-02-17 | 1.39 | Fixed Library search feature ‘Use max dRi’ (Method – Library List) |
| 2025-02-19 | 1.40 | Improved Spectrum view with colors and ctrl+leftclick on cas |
| 2025-03-11 | 1.41 | Fixed Spectrum panning crash |
| 2025-04-26 | 1.42 | Report Template with VBA macro functionality to produce chromatogram in Excel graphics |
| 2025-05-09 | 1.43 | Lineup export with Pivot template and chromatogram |
| 2025-06-23 | 1.44 | Excel XLMX Report cleaned properly before creating the report. Cosmetic. |
| 2026-01-12 | 1.47 | Skipped version numbers. Added new features to Lineup view. |
| 2026-01-21 | 1.48 | Added signal to noise calculations |
| 2026-01-23 | 1.49 | Deconvolution signal noise calculation >10x faster |
| 2026-01-27 | 1.50 | Improved IonChromatogram window performance |